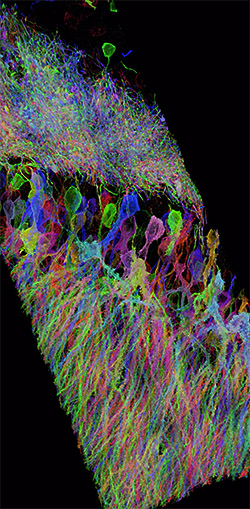
Expansion microscopy (ExM) is a method for improving the resolution of light
microscopy by physically expanding biological samples.
It allows investigators to identify small structures in 3-D and with Nano-scale
resolution by expanding them with a polymer system.
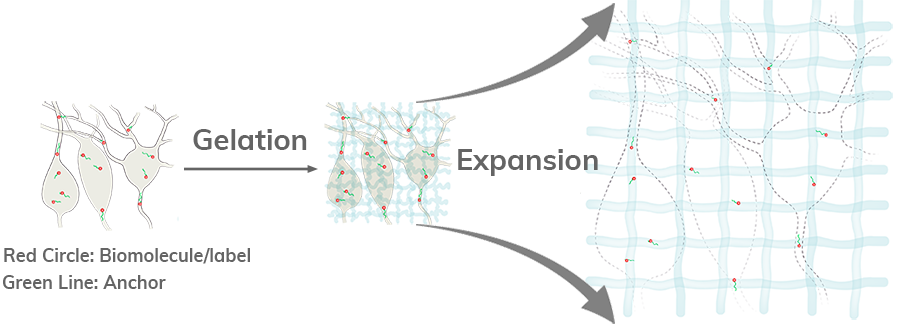
In ExM, biomolecules and/or fluorescent labels in the specimen are linked to a
dense, expandable polymer matrix synthesized evenly throughout the specimen, which
undergoes 3-dimensional expansion by about 4.5 fold linearly when immersed in water
[1].
A recent paper reports that multiple applications of ExM expands biological
specimens up to 20 fold, and enables ~25 nm resolution imaging of cells and tissues
on conventional microscopes [2].
Traditional light microscopy has limits of resolution that prevent it from reliably
distinguishing small structures that are important for biological function.
For example, synaptic vesicles are 40-50 nano meters in diameter, which is below
the
commonly quoted resolution limit of 200 nano meters required for light microscopy.
There are a few methods to archive the nano meter scale image, such as Super
Resolution microscopy and Electron microscopy, but these methods require special
hardware that is complex and/or expensive. In addition, they are physically limited
in imaging speed, number of colors, and/or in the volume accessible, as compared to
diffraction-limited microscopy. ExM enables the nano scale resolution imaging with
standard diffraction-limited microscope hardware already common in biology labs.
Moreover, since ExM expands samples in water, final expanded specimen is ~99%
water, and thus are essentially completely transparent and optical aberration free
[3].
Another benefit of ExM is the decrowding of biomolecules or labels by expanding
them away from each other. This allows for more room around the molecules for
chemical
reactions to take place.
This extra room can be used to perform signal amplification, so that rather than
attempting difficult single molecule imaging in 3-D volumes of intact tissue, one
can attach many fluorophores to a single-molecule biomolecular target. Which will
allow the
identification and localization of single biomolecules feasible in large volumes
(as demonstrated for this method applied to the detection of single RNAs in
synapses in intact brain tissue in [4], and shown in Fig. 2f, g).
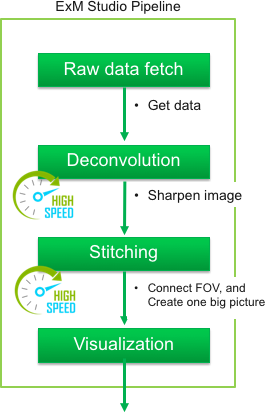
The purpose of this web site is to provide the information and tools for performing
ExM
procedures to every bioscientist familiar with immunohistochemistry.